BACK END/R
[R] R 정리 15 - Decision Tree
circle kim
2021. 2. 2. 10:55
# 분류모델 중 Decision Tree
set.seed(123)
ind <- sample(1:nrow(iris), nrow(iris) * 0.7, replace = FALSE)
train <- iris[ind, ]
test <- iris[-ind, ]
# ctree
install.packages("party")
library(party)
iris_ctree <- ctree(formula = Species ~ ., data = train)
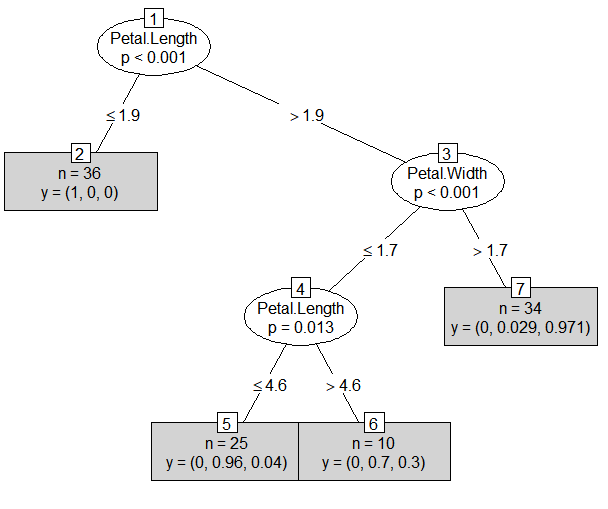
iris_ctree
# Conditional inference tree with 4 terminal nodes
#
# Response: Species
# Inputs: Sepal.Length, Sepal.Width, Petal.Length, Petal.Width
# Number of observations: 105
#
# 1) Petal.Length <= 1.9; criterion = 1, statistic = 97.466
# 2)* weights = 36
# 1) Petal.Length > 1.9
# 3) Petal.Width <= 1.7; criterion = 1, statistic = 45.022
# 4) Petal.Length <= 4.6; criterion = 0.987, statistic = 8.721
# 5)* weights = 25
# 4) Petal.Length > 4.6
# 6)* weights = 10
# 3) Petal.Width > 1.7
# 7)* weights = 34
plot(iris_ctree, type = "simple")
plot(iris_ctree)

# predit
pred <- predict(iris_ctree, test)
pred
t <- table(pred, test$Species) # 인자 : 예측값, 실제값
t
# pred setosa versicolor virginica
# setosa 14 0 0
# versicolor 0 18 1
# virginica 0 0 12
sum(diag(t)) / nrow(test) # 0.9777778library(caret)
confusionMatrix(pred, test$Species)
# Confusion Matrix and Statistics
#
# Reference
# Prediction setosa versicolor virginica
# setosa 14 0 0
# versicolor 0 18 1
# virginica 0 0 12
#
# Overall Statistics
#
# Accuracy : 0.9778
# 95% CI : (0.8823, 0.9994)
# No Information Rate : 0.4
# P-Value [Acc > NIR] : < 2.2e-16
#
# Kappa : 0.9662
#
# Mcnemar's Test P-Value : NA
#
# Statistics by Class:
#
# Class: setosa Class: versicolor Class: virginica
# Sensitivity 1.0000 1.0000 0.9231
# Specificity 1.0000 0.9630 1.0000
# Pos Pred Value 1.0000 0.9474 1.0000
# Neg Pred Value 1.0000 1.0000 0.9697
# Prevalence 0.3111 0.4000 0.2889
# Detection Rate 0.3111 0.4000 0.2667
# Detection Prevalence 0.3111 0.4222 0.2667
# Balanced Accuracy 1.0000 0.9815 0.9615
# 방법2 : rpart : 가지치기 이용
library(rpart)
iris_rpart <- rpart(Species ~ ., data = train, method = 'class')
x11()
plot(iris_rpart)
text(iris_rpart)
plotcp(iris_rpart)
printcp(iris_rpart)

cp <- iris_rpart$cptable[which.min(iris_rpart$cptable[, 'xerror'])]
iris_rpart_prune <- prune(iris_rpart, cp=cp, 'cp')
x11()
plot(iris_rpart_prune)
text(iris_rpart_prune)